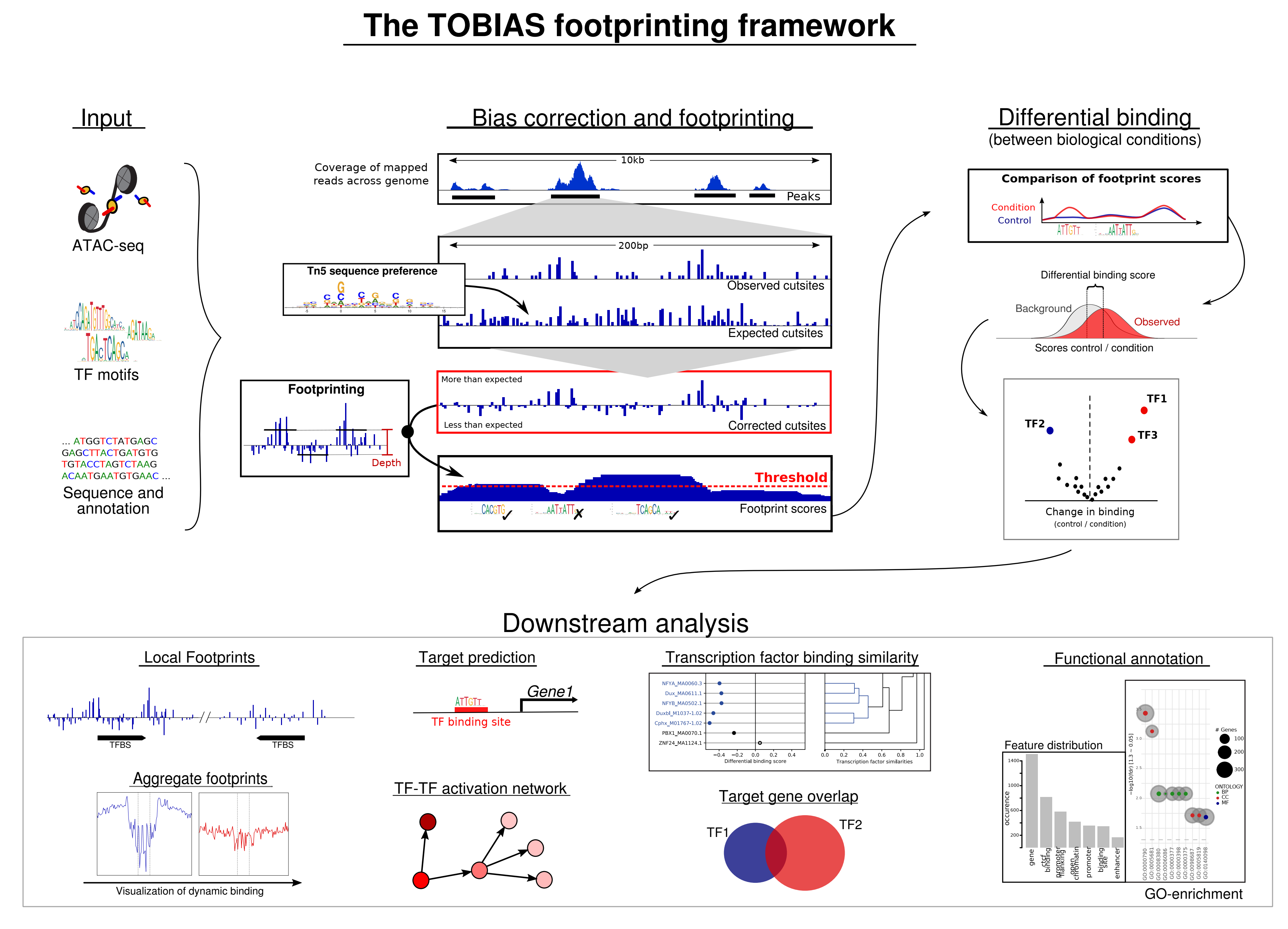
A complete framework for performing footprinting analysis on ATAC-seq data
TOBIAS
Main features
- All in one digital genomic footprinting framework
- Easy to use
- Powerfull downstream analysis modules
- Universal file formats
ATAC-seq (Assay for Transposase-Accessible Chromatin using high-throughput sequencing) is a sequencing assay for investigating genome-wide chromatin accessibility. The assay applies a Tn5 Transposase to insert sequencing adapters into accessible chromatin, enabling mapping of regulatory regions across the genome. Additionally, the local distribution of Tn5 insertions contains information about transcription factor binding due to the visible depletion of insertions around sites bound by protein - known as footprints.
TOBIAS integrates these footprints with genomic information and transcription factor motifs to predict transcription factor binding. TOBIAS is collection of command tools written in Python, with each tool is intended to be used in a framework approach as illustrated below: