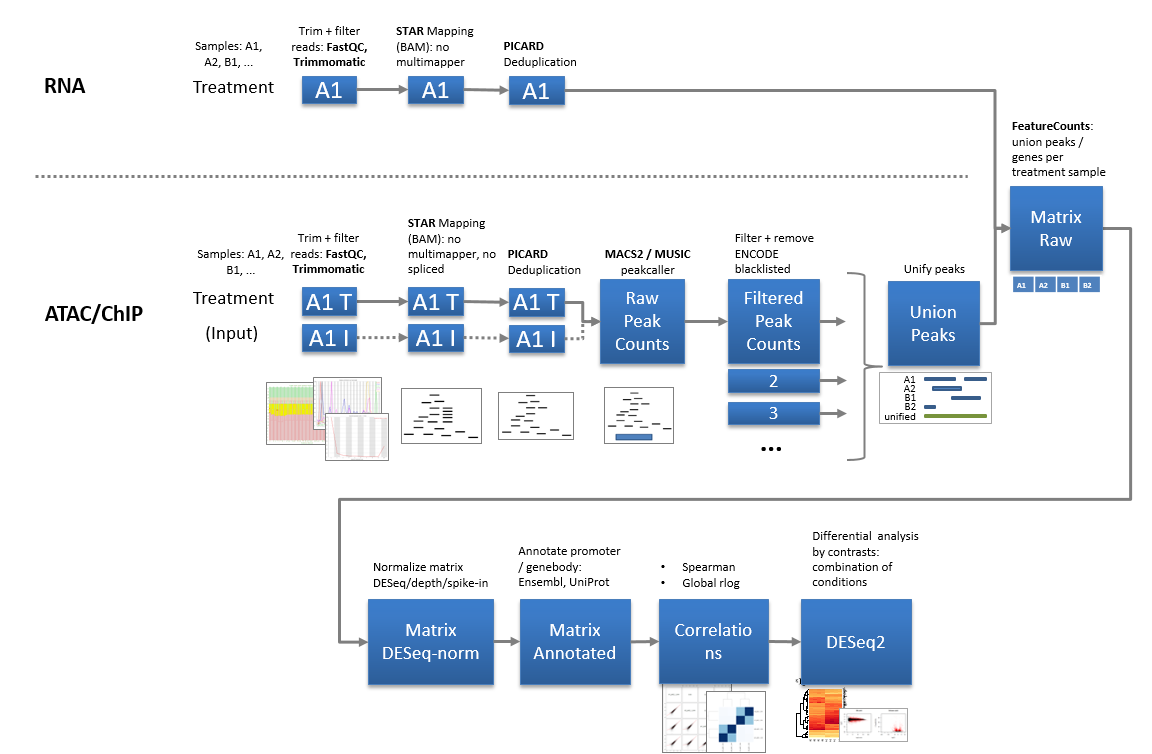
Pipelines to process bulk high throughput sequencing data.
Bulk Pipelines
Bulk pipelines are based on mapping/counting reads at known features (e.g. genes) or at loci with significant enrichment (peaks). Typical steps include QC, differential expression analyses, annotation, clustering, and gene set enrichment analyses. Pipelines are heavily integrated with internal hard- and software resources and unfortunately not runnable externally.