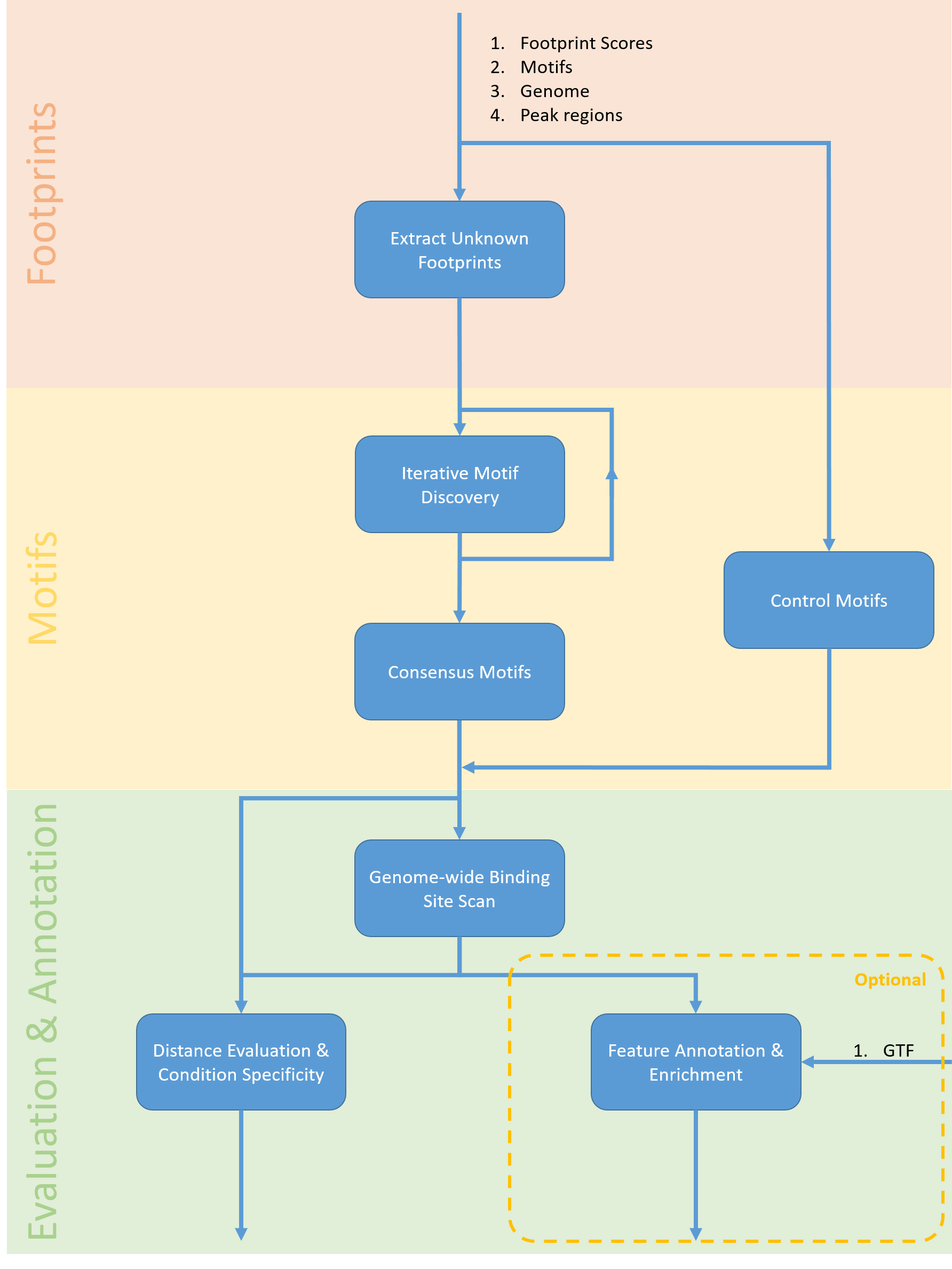
A pipeline for generation of novel transcription factor motifs
DENIS
ATAC-seq (Assay for Transposase Accessible Chromatin using sequencing) allows for the genome-wide assessment of chromatin accessibility. This is done by a hyperactive Tn5 transposase, which cuts open chromatin regions of the DNA into fragments. After amplification, sequencing, and analysis of the ATAC-seq data, open chromatin regions are identified as an accumulation of reads, named as peaks. A closer look at these peaks reveals so-called footprints, defined as small regions of less read coverage that indicate DNA-protein binding. So far, footprints have exclusively been assigned to known motifs, enabling an important but incomplete understanding on TF roles in networks. However, since the existence of a footprint implies binding of a protein and further provides information on binding preference, it can be assumed that footprints without binding motifs code for previously undetected motifs.
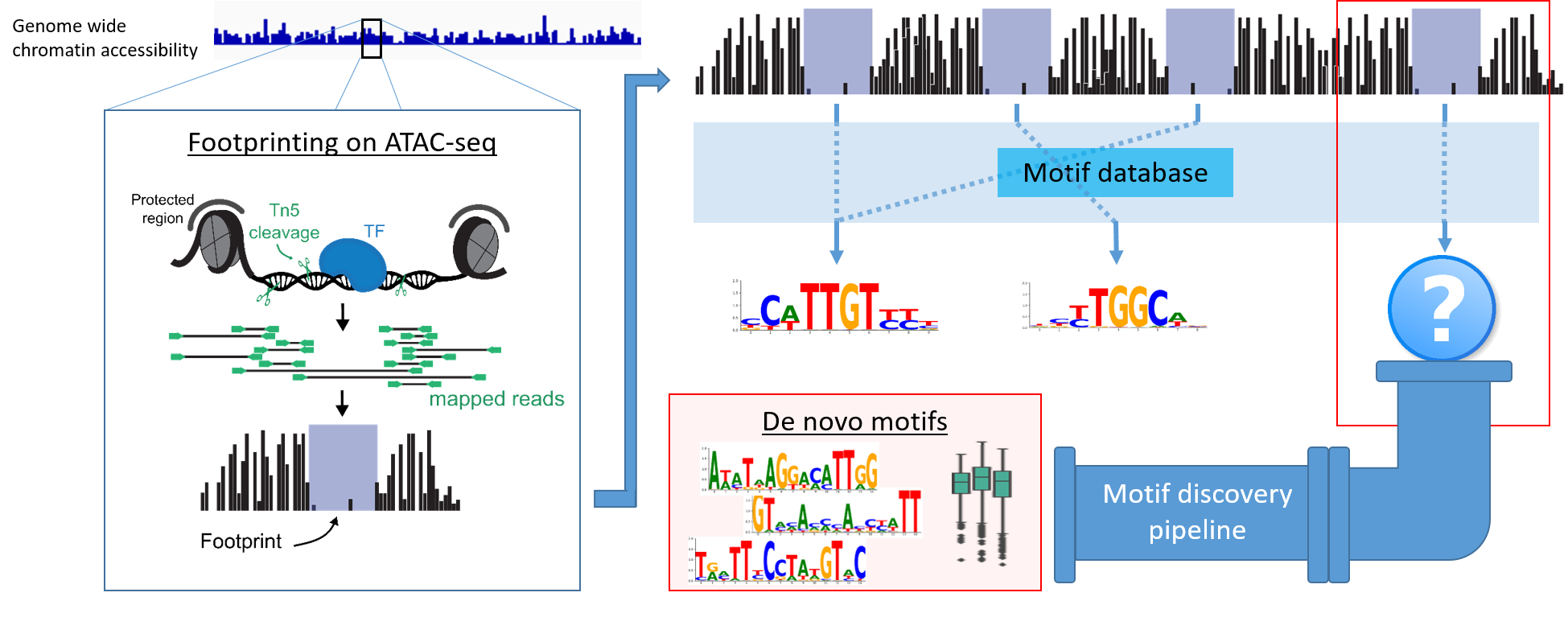
In order to systematically investigate the potential of an computational approach of de novo motif discovery based on footprints, a pipeline was implemented that i) isolates footprints without binding motifs, ii) performs de novo motif generation and iii) validates these motifs through enrichment analysis and annotation. As a proof of principle the pipeline is shown to rediscover artificially removed motifs and an exemplary downstream analysis on a wildtype/TF-overexpression dataset reveals differentially regulated de novo motifs.