Our software
robust proportion analysis for single cell resolution data
Scanpro
 Scanpro is a modular tool for proportion analysis, seamlessly integrating into widely accepted frameworks in the python environment. Scanpro is fast, accurate, support datasets without replicates, and is intended to be used by bioinformatics experts and beginners. read more
Scanpro is a modular tool for proportion analysis, seamlessly integrating into widely accepted frameworks in the python environment. Scanpro is fast, accurate, support datasets without replicates, and is intended to be used by bioinformatics experts and beginners. read more
A framework to streamline single cell analysis.
SC-Framework
 The SC-Framework is comprised of a python package and a collection of jupyter notebooks to enable a concise and reproducible method to do single cell analysis. read more
The SC-Framework is comprised of a python package and a collection of jupyter notebooks to enable a concise and reproducible method to do single cell analysis. read more
Nucleosome Positioning - NucleoDetective.
NucleoDetective
 NucleoDetective is a tool to predict nucleosome positions using ATAC-seq data and to compare the found patterns across different conditions. read more
NucleoDetective is a tool to predict nucleosome positions using ATAC-seq data and to compare the found patterns across different conditions. read more
Pipelines to process bulk high throughput sequencing data.
Bulk Pipelines
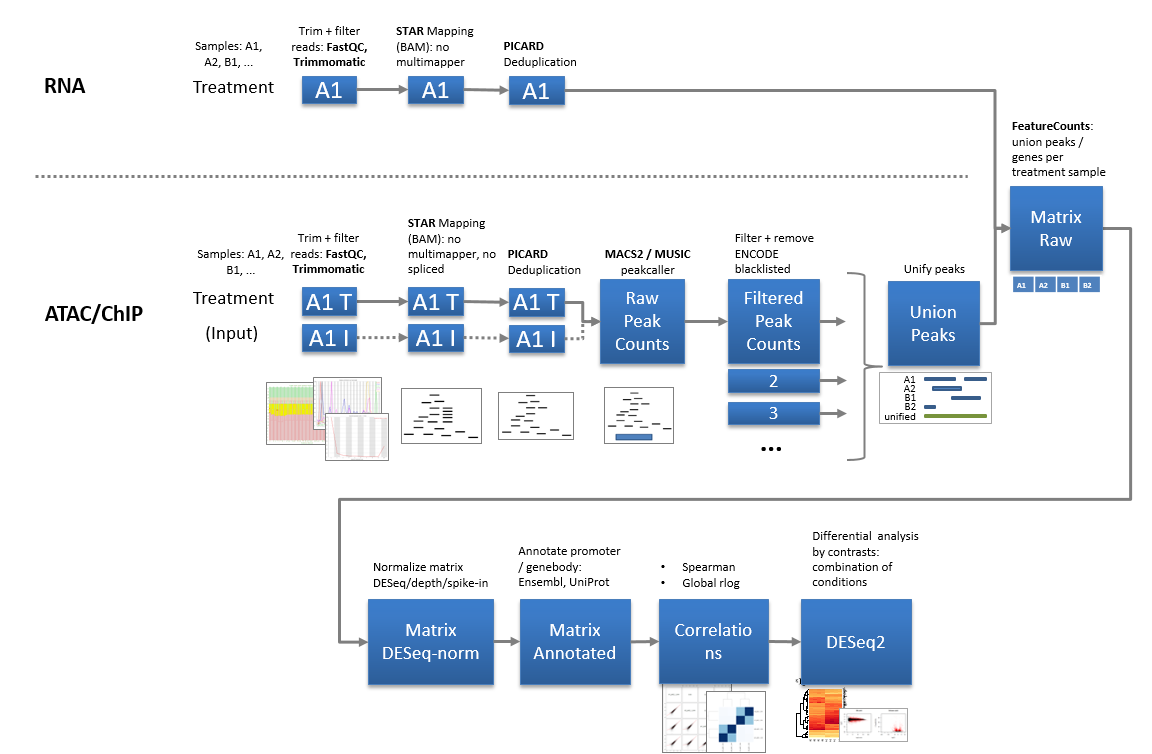 Bulk RNA/ATAC/ChIP-Seq are analyzed via in-house software pipelines. read more
Bulk RNA/ATAC/ChIP-Seq are analyzed via in-house software pipelines. read more
Find single nucleotide polymorphisms for precise ORF disruption
WobbleDORF
 WobbleDORF is an modular structured command line tool to find single nucleotide polymorphisms, that would disrupt an open reading frame. Main Feature is the frame specific preservation via wobble bases. read more
WobbleDORF is an modular structured command line tool to find single nucleotide polymorphisms, that would disrupt an open reading frame. Main Feature is the frame specific preservation via wobble bases. read more
Design and implementation of a tool to identify genetic directional dependencies.
Genetic directional dependencies
 Prediction of combinatorial phenotypes in which the order of mutations direct the phenotype from CRISPR pertubation screens. read more
Prediction of combinatorial phenotypes in which the order of mutations direct the phenotype from CRISPR pertubation screens. read more
A R based package for gRNA design
multicrispr²
 multicrispr² is a R based pacakge for designing gRNAs with the special feature of designing gRNA pairs intended for genomic excision. read more
multicrispr² is a R based pacakge for designing gRNAs with the special feature of designing gRNA pairs intended for genomic excision. read more
MAnaging Computing SErvices on Kubernetes
MACSEK
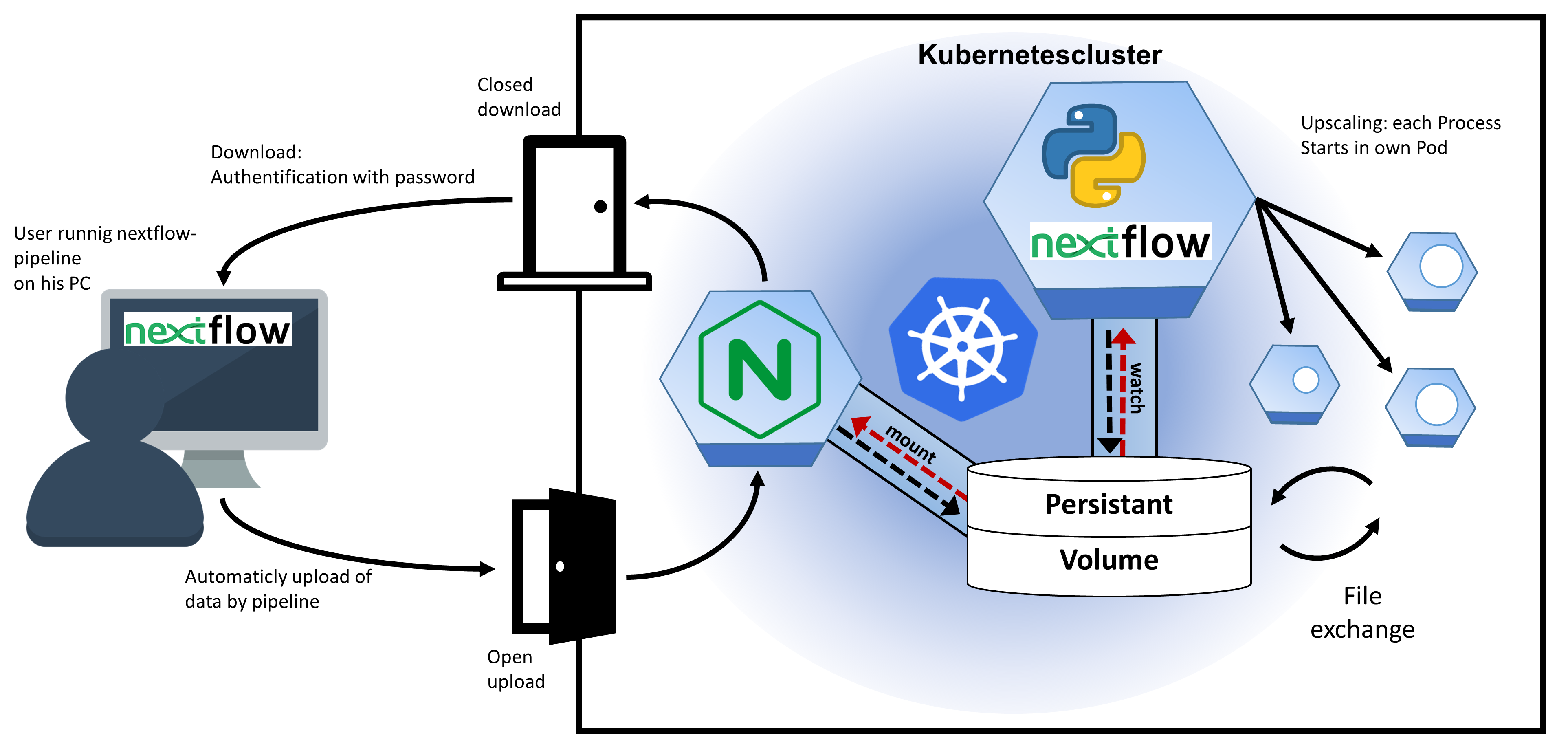 MACSEK(MAnaging Computing SErvices on Kubernetes) is a framework that provides a computing service on Kubernetes cluster, such as provided by deNBi. It creates a service on the clusters, which allows the upload of files for all kinds of calculation, manages resource allocation on the cluster and provides the download of the results. read more
MACSEK(MAnaging Computing SErvices on Kubernetes) is a framework that provides a computing service on Kubernetes cluster, such as provided by deNBi. It creates a service on the clusters, which allows the upload of files for all kinds of calculation, manages resource allocation on the cluster and provides the download of the results. read more
Iterative & Interactive dashboards
i2dash
 Scientific communication and data visualization are important aspects to illustrate complex concepts and results from data analyses. The R package i2dash provides functionality to create customized, web-based dashboards for data presentation, exploration and sharing. i2dash integrates easily into existing data analysis pipelines and can organize scientific findings thematically across different pages and layouts. read more
Scientific communication and data visualization are important aspects to illustrate complex concepts and results from data analyses. The R package i2dash provides functionality to create customized, web-based dashboards for data presentation, exploration and sharing. i2dash integrates easily into existing data analysis pipelines and can organize scientific findings thematically across different pages and layouts. read more
MAnaging Multiple Projects On Kubernetes
MaMPOK
 MaMPOK is an offline Kubernetes management tool. It is intented to use Kubernetes cloud computing at de.NBi. It provides methods to deploy e.g. web apps as containers automatically. It automates file transfers via a S3 object storage, takes care of container images, and keep track of a large number of projects. read more
MaMPOK is an offline Kubernetes management tool. It is intented to use Kubernetes cloud computing at de.NBi. It provides methods to deploy e.g. web apps as containers automatically. It automates file transfers via a S3 object storage, takes care of container images, and keep track of a large number of projects. read more
Visualization of omics data
WIlsON
 High-throughput (HT) studies of complex biological systems generate a massive amount of omics data. The results are typically summarized using spreadsheet like file formats. Visualization of this data is a key aspect of both, the analysis and the understanding of biological systems under investigation. While users have many visualization methods and tools to choose from, the challenge is to properly handle these tools and create clear, meaningful, and integrated visualizations based on pre-processed datasets. read more
High-throughput (HT) studies of complex biological systems generate a massive amount of omics data. The results are typically summarized using spreadsheet like file formats. Visualization of this data is a key aspect of both, the analysis and the understanding of biological systems under investigation. While users have many visualization methods and tools to choose from, the challenge is to properly handle these tools and create clear, meaningful, and integrated visualizations based on pre-processed datasets. read more
a tool for Universal RObust Peak Annotation
UROPA
 UROPA (Universal RObust Peak Annotator) is a command line based tool, intended for universal genomic range annotation. Based on a configuration file, different target features can be prioritized with multiple integrated queries. These can be sensitive for feature type, distance, strand specificity, feature attributes (e.g. protein_coding) or anchor position relative to the feature. read more
UROPA (Universal RObust Peak Annotator) is a command line based tool, intended for universal genomic range annotation. Based on a configuration file, different target features can be prioritized with multiple integrated queries. These can be sensitive for feature type, distance, strand specificity, feature attributes (e.g. protein_coding) or anchor position relative to the feature. read more
Analysis and visualization of differential methylation in genomic regions
ADMIRE
 DNA methylation at cytosine nucleotides constitutes epigenetic gene regulation impacting cellular development and the stage of a disease. Besides whole genome bisulfit sequencing, Illumina HumanMethylationEPIC Assays represent a versatile and cost-effective tool to investigate changes of methylation patterns at CpG sites. ADMIRE is a semi-automatic analysis pipeline and visualization tool for Infinium HumanMethylation450K and Infinium MethylationEpic assays. read more
DNA methylation at cytosine nucleotides constitutes epigenetic gene regulation impacting cellular development and the stage of a disease. Besides whole genome bisulfit sequencing, Illumina HumanMethylationEPIC Assays represent a versatile and cost-effective tool to investigate changes of methylation patterns at CpG sites. ADMIRE is a semi-automatic analysis pipeline and visualization tool for Infinium HumanMethylation450K and Infinium MethylationEpic assays. read more