-
Notifications
You must be signed in to change notification settings - Fork 0
Advanced usage
WIlsON is primarily intended to empower screening platforms to offer access to pre-calculated HT screen results to the non-computational scientist.
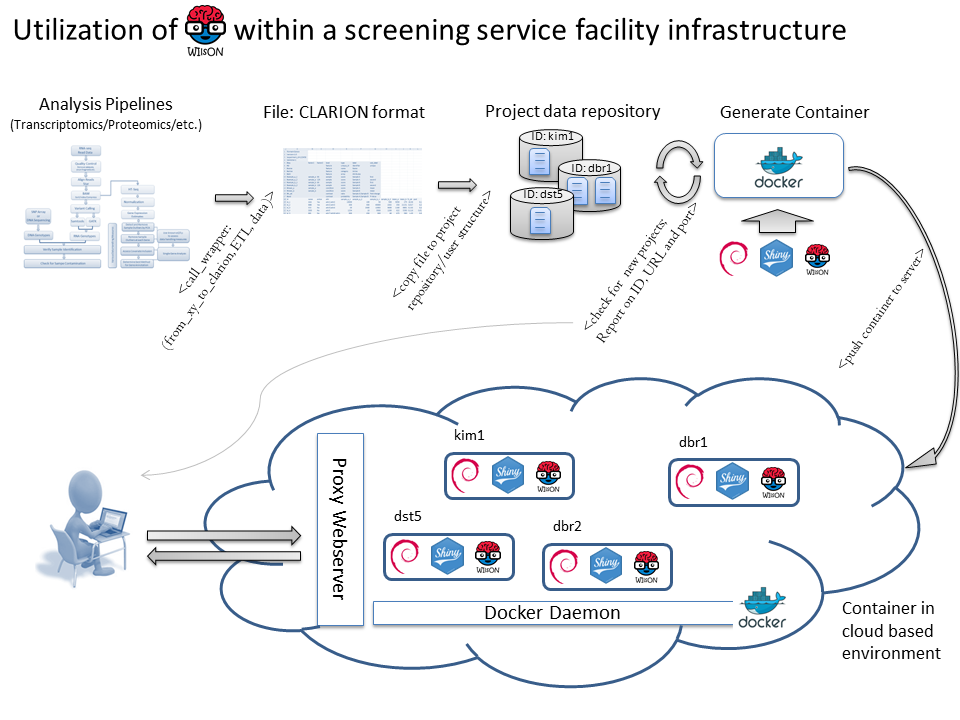
Thereby, a screening platform generates raw data that is analyzed by a platform-specific software pipeline, providing a platform-specific result format. An ETL (Extract, Transform, Load) process extracts relevant data and generates a CLARION file, that is copied to a folder structure reflection groups/experiments/projects. A cronjob checks the folder structure and generates new containers for each newly established project. Containers are started in a cloud infrastructure running a docker daemon and webproxy server that is aware of all individual containers. In addition, the container script randomly generates URLs and ports that are send to the user and webproxy for container administration.
How to cite?
H. Schultheis, C. Kuenne, J. Preussner, R. Wiegandt, A. Fust, M. Bentsen and M. Looso. WIlsON: Webbased Interactive Omics VisualizatioN. Bioinformatics 35(6) 2018, doi: https://doi.org/10.1093/bioinformatics/bty711
Copyright © 2019 Dr. Mario Looso, Max Planck Institute for Heart and Lung Research